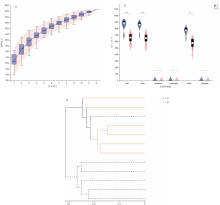
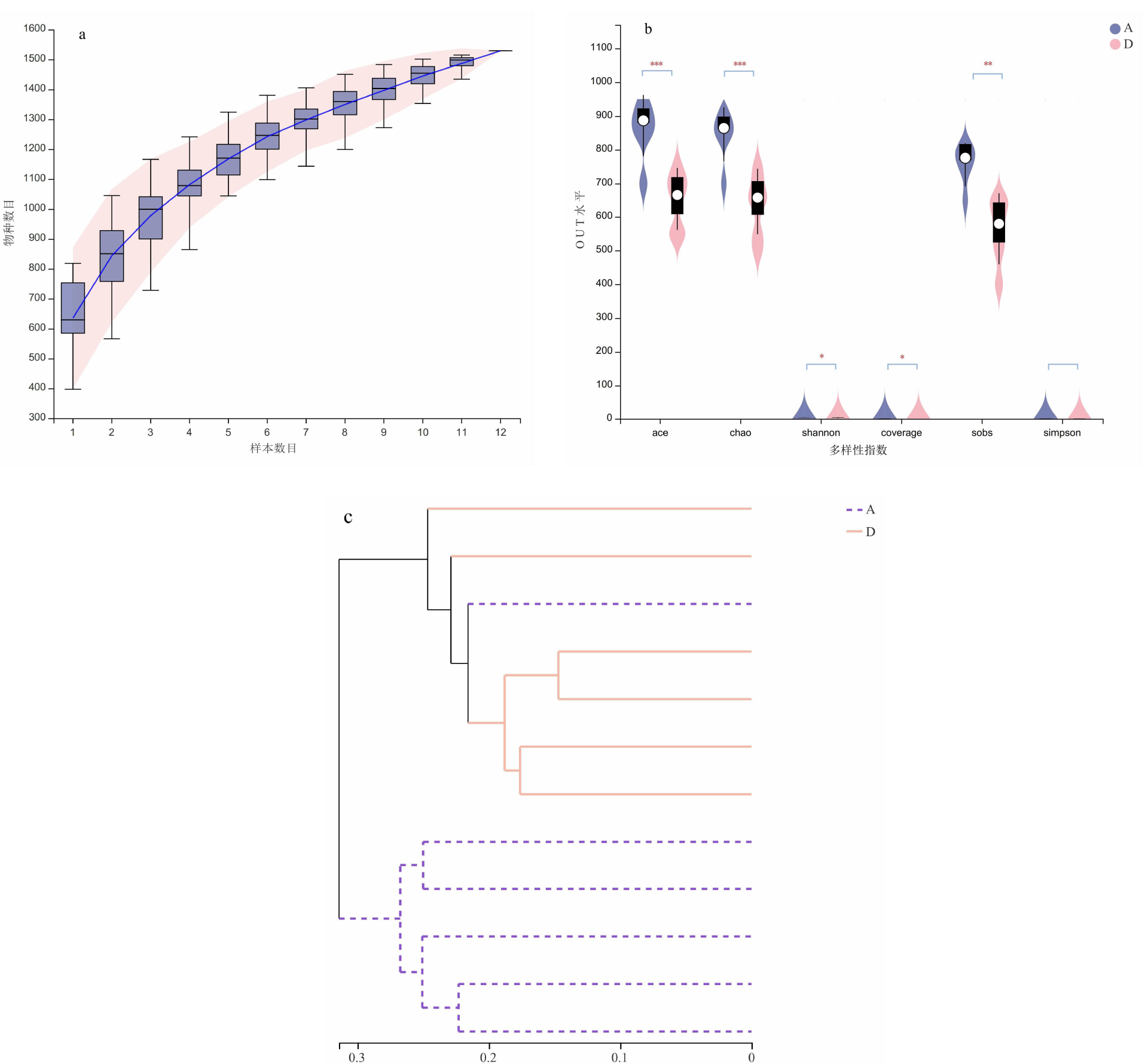
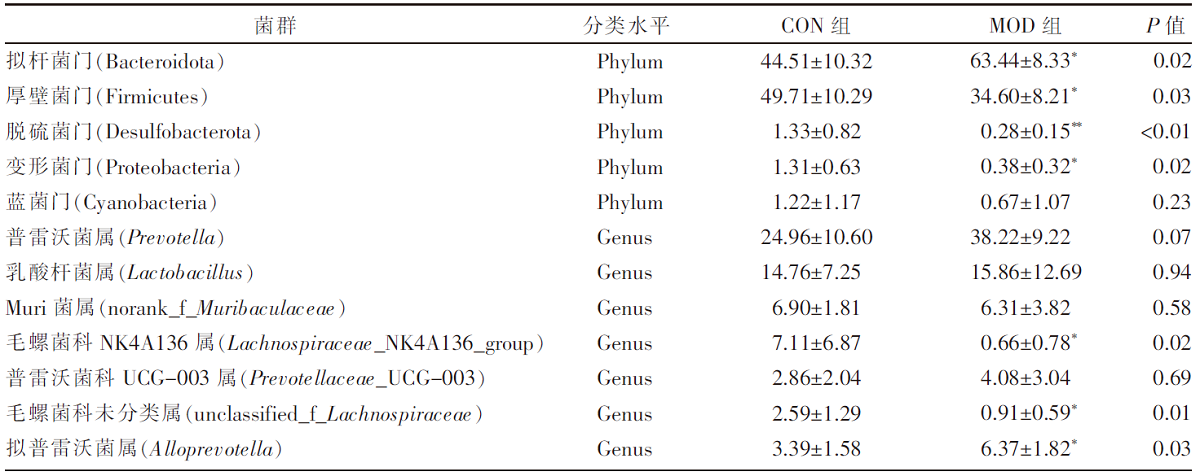
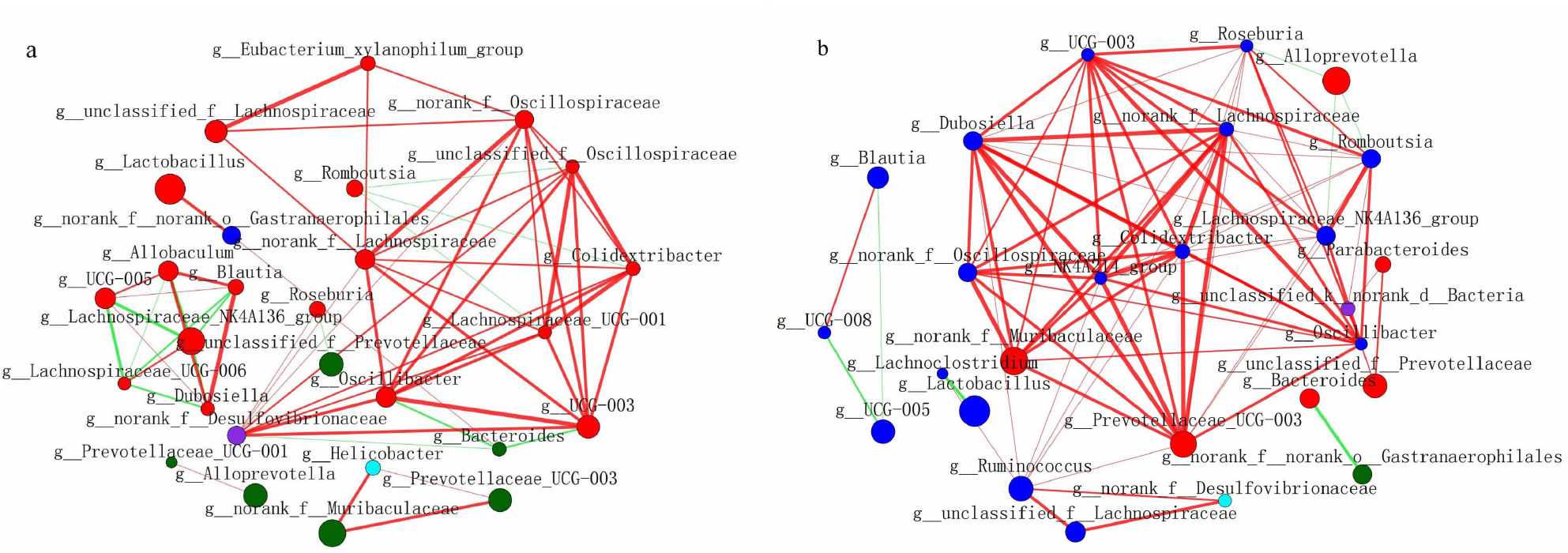
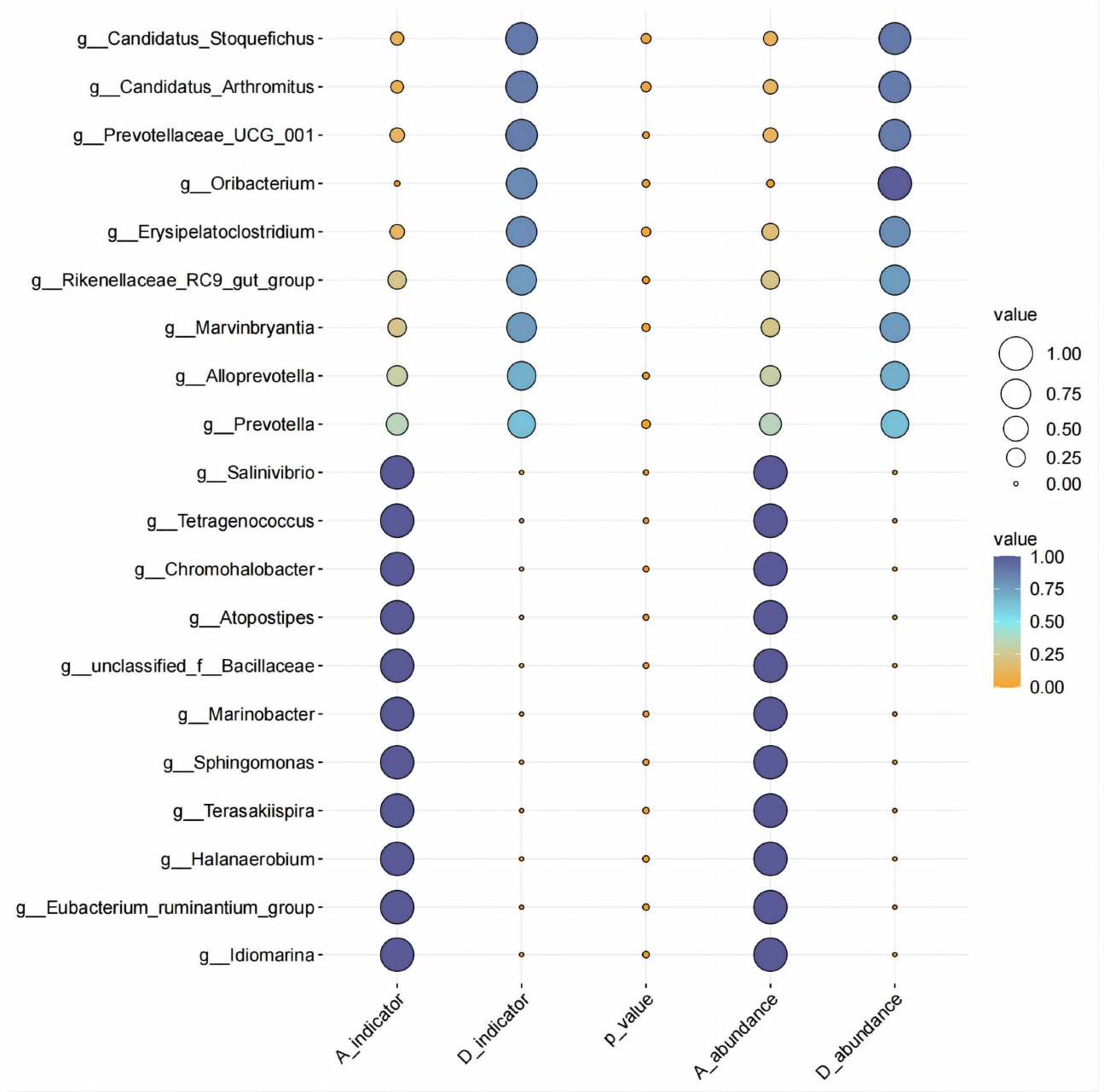
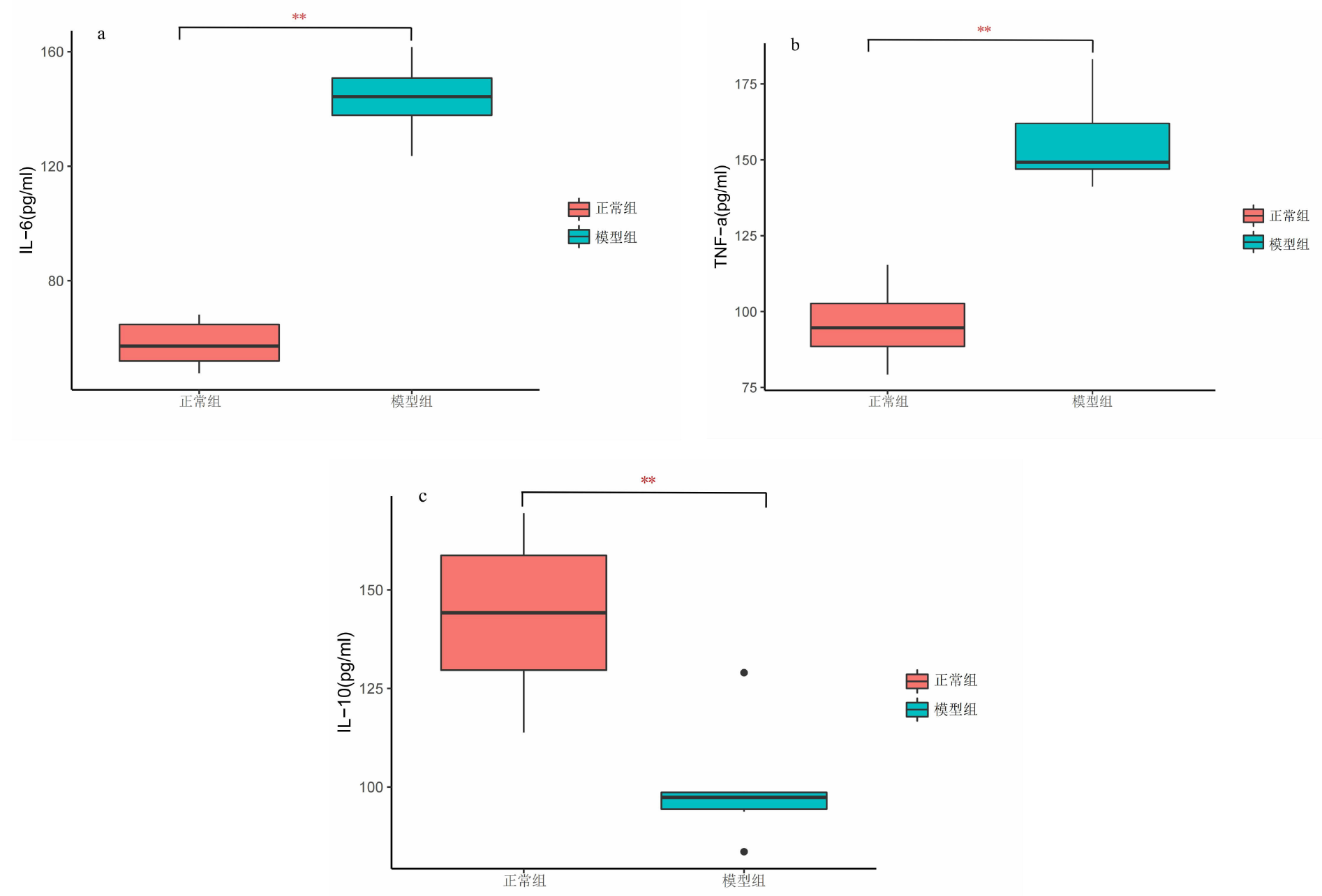
| [1] |
BASHASHATI M, REZAEI N, SHAFIEYOUN A, et al. Cytokine imbalance in irritable bowel syndrome: A systematic review and meta-analysis[J]. Neurogastroenterology and Motility, 2014, 26(7): 1036-1048.
doi: 10.1111/nmo.12358
pmid: 24796536
|
| [2] |
FORD A C, SPERBER A D, CORSETTI M, et al. Irritable bowel syndrome[J]. The Lancet, 2020, 396(10263):1675-1688.
doi: 10.1016/S0140-6736(20)31548-8
|
| [3] |
BERTRAND J, MARION-LETELLIER R, AZHAR S, et al. Glutamine enema regulates colonic ubiquitinated proteins but not proteasome activities during TNBS-induced colitis leading to increased mitochondrial activity[J]. Proteomics, 2015, 15(13):2198-2210.
doi: 10.1002/pmic.201400304
pmid: 25689466
|
| [4] |
BIANCONE L, ARDIZZONE S, ARMUZZI A, et al. Ustekinumab for treating ulcerative colitis: An expert opinion[J]. Expert Opinion on Biological Therapy, 2020, 20(11):1321-1329.
doi: 10.1080/14712598.2020.1792882
|
| [5] |
BORRIELLO F, POLI V, SHROCK E, et al. An adjuvant strategy enabled by modulation of the physical properties of microbial ligands expands antigen immunogenicity[J]. Cell, 2022, 185(4):614-629.
doi: 10.1016/j.cell.2022.01.009
pmid: 35148840
|
| [6] |
张儒奇, 方志安, 韩文庆, 等. 葛根芩连汤对腹泻型肠易激综合征大鼠肠道菌群的影响[J]. 中国中药杂志, 2022, 47(24):6709-6719.
|
| [7] |
CHEN Y, XIAO S, GONG Z, et al. Wuji Wan formula ameliorates diarrhea and disordered colonic motility in post-inflammation irritable bowel syndrome rats by modulating the gut microbiota[J]. Frontiers in Microbiology, 2017, 8:2307.
doi: 10.3389/fmicb.2017.02307
pmid: 29218037
|
| [8] |
郑璐玉. 痰湿体质人群炎症相关机制研究[D]. 北京: 北京中医药大学, 2013.
|
| [9] |
李榕娇, 郭维龙, 孙慧冰, 等. 肠易激综合征患者肠道菌群分布和炎症因子及血清NPY、SP、5-HT水平的变化[J]. 中国微生态学杂志, 2020, 32(9):1060-1064.
|
| [10] |
贺星. TLR4/NF-κB信号通路在腹泻型肠易激综合征中的作用及机制研究[D]. 北京: 中国人民解放军陆军军医大学, 2018.
|
| [11] |
赵斌, 郭联斌, 段武琼. 肠易激综合征血清炎症因子表达与肠道菌群变化的关系[J]. 现代消化及介入诊疗, 2019, 24(9):1046-1049.
|
| [12] |
BROWN R L, LARKINSON M L Y, CLARKE T B. Immunological design of commensal communities to treat intestinal infection and inflammation[J]. PLoS Pathogens, 2021, 17(1):e1009191.
|
| [13] |
ILJAZOVIC A, ROY U, GÁLVEZ E J C, et al. Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation[J]. Mucosal Immunology, 2021, 14(1):113-124.
doi: 10.1038/s41385-020-0296-4
|
| [14] |
TAMANAI-SHACOORI Z, SMIDA I, BOUSARGHIN L, et al. Roseburia spp.: A marker of health?[J]. Future Microbiology, 2017, 12(2):157-170.
doi: 10.2217/fmb-2016-0130
|
| [15] |
殷瑞霞, 皇甫建, 肖瑞, 等. 2型糖尿病及并发肾病模型大鼠肠道菌群多样性分析[J]. 动物医学进展, 2022, 43(4):30-37.
|
| [16] |
丁姮月, 朱惠萍, 梁国强, 等. 基于高通量测序技术的脾虚腹泻型肠易激综合征小鼠与健康小鼠肠道菌群的差异研究[J]. 中国中医基础医学杂志, 2021, 27(2):260-266,324.
|
| [17] |
HEDBLOM G A, REILAND H A, SYLTE M J, et al. Segmented filamentous bacteria-metabolism meets immunity[J]. Frontiers in Microbiology, 2018, 9:1991.
doi: 10.3389/fmicb.2018.01991
|
| [18] |
LI A L, NI W W, ZHANG Q M, et al. Effect of cinnamon essential oil on gut microbiota in the mouse model of dextran sodium sulfate-induced colitis[J]. Microbiology and Immunology, 2020, 64(1):23-32.
doi: 10.1111/mim.v64.1
|
| [19] |
SHI K, QU L H, LIN X, et al. Deep-fried atractylodis rhizoma protects against spleen deficiency-induced diarrhea through regulating intestinal inflammatory response and gut microbiota[J]. International Journal of Molecular Sciences, 2019, 21(1): 124.
doi: 10.3390/ijms21010124
|
| [20] |
冯荣建, 余茜, 李怡, 等. 卒中后认知功能障碍患者肠道菌群紊乱特征研究[J]. 四川大学学报(医学版), 2021, 52(6):966-974.
|