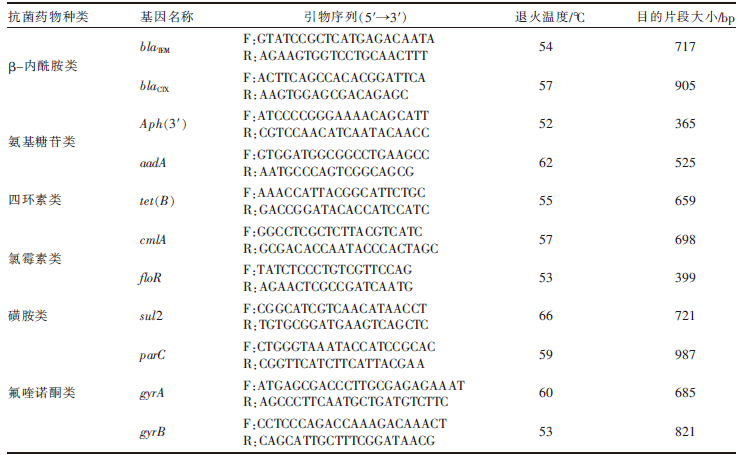
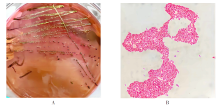
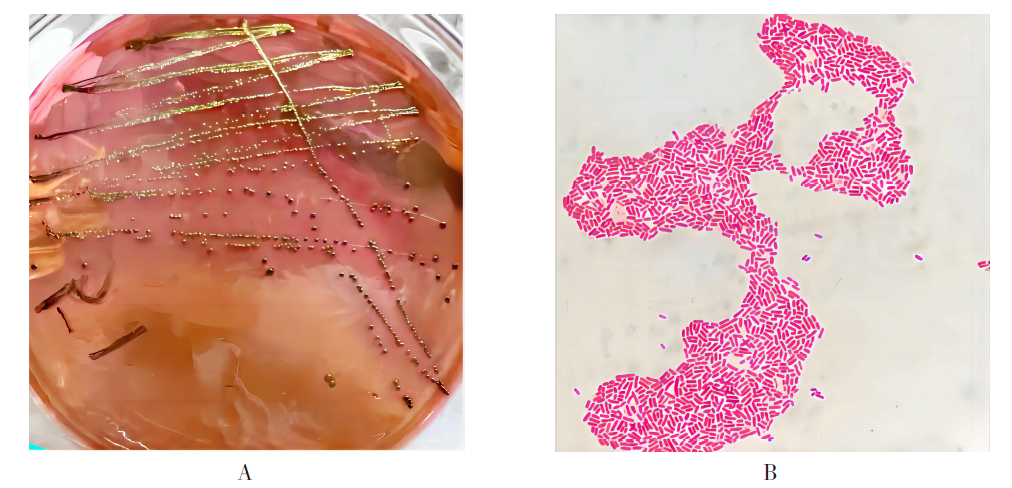
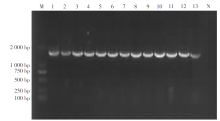
| [1] |
吕玉金. 致脑膜炎大肠杆菌感染脑微血管内皮细胞的蛋白质组学研究[D]. 武汉: 华中农业大学, 2019.
|
| [2] |
DERAKHSHAN S, FARHADIFAR F, ROSHANI D, et al. Study on the presence of resistant diarrheagenic pathotypes in Escherichia coli isolated from patients with urinary tract infection[J]. Gastroenterology and Hepatology from Bed to Bench, 2019, 12(4):348-357.
|
| [3] |
李宏博. 伊犁昭苏不同动物源大肠杆菌耐药性检测及相关耐药基因分析[D]. 乌鲁木齐: 新疆农业大学, 2022.
|
| [4] |
左勇, 李建玲, 王涛, 等. 马大肠杆菌感染的诊治和预防[J]. 当代畜牧, 2021(11):24-25.
|
| [5] |
KENNEDY C A, WALSH C, KARCZMARCZYK M, et al. Multi-drug resistant Escherichia coli in diarrhoeagenic foals:Pulsotyping, phylotyping, serotyping, antibiotic resistance and virulence profiling[J]. Veterinary Microbiology, 2018, 223:144-152.
doi: 10.1016/j.vetmic.2018.08.009
|
| [6] |
姚火春. 兽医微生物学实验指导[M]. 2版. 北京: 中国农业出版社, 2002.
|
| [7] |
杨滴, 王耀兵, 李冬梅, 等. 粪便中大肠埃希菌的分离鉴定[J]. 微生物学杂志, 2007, 27(3):1-5.
|
| [8] |
CLERMONT O, BONACORSI S, BINGEN E. Rapid and simple determination of the Escherichia coli phylogenetic group[J]. Applied and Environmental Microbiology, 2000, 66(10):4555-4558.
doi: 10.1128/AEM.66.10.4555-4558.2000
|
| [9] |
胡会杰, 张琪, 周明旭, 等. 不同禽源致病性大肠杆菌毒力基因分布规律研究[J]. 中国家禽, 2015, 37(10):34-37.
|
| [10] |
刘英玉, 朱明月, 苏晓月, 等. 新疆库尔勒地区羊源产志贺毒素大肠杆菌及其耐药性分析[J]. 中国农业科技导报, 2020, 22(5):122-128.
doi: 10.13304/j.nykjdb.2019.0053
|
| [11] |
刘少昆. 京津部分地区犊牛腹泻大肠杆菌分离鉴定及耐药性分析[D]. 长春: 吉林农业大学, 2016.
|
| [12] |
YILMAZ E S, ASLANTAS Ö. Phylogenetic group/subgroups distributions, virulence factors, and antimicrobial susceptibility of Escherichia coli strains from urinary tract infections in Hatay[J]. Revista Da Sociedade Brasileira De Medicina Tropical, 2020, 53:e20190429.
doi: 10.1590/0037-8682-0429-2019
|
| [13] |
CLERMONT O, CHRISTENSON J K, DENAMUR E, et al. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups[J]. Environmental Microbiology Reports, 2013, 5(1):58-65.
doi: 10.1111/1758-2229.12019
pmid: 23757131
|
| [14] |
江婉琳, 徐双军, 刘雨欣, 等. 新疆阿克苏地区奶牛粪源大肠杆菌分型与耐药性分析[J]. 中国兽医学报, 2022, 42(12):2434-2443.
|
| [15] |
CHUNG Y S, SONG J W, KIM D H, et al. Isolation and characterization of antimicrobial-resistant Escherichia coli from national horse racetracks and private horse-riding courses in Korea[J]. Journal of Veterinary Science, 2016, 17(2):199-206.
doi: 10.4142/jvs.2016.17.2.199
|
| [16] |
佟盼盼, 黄顺敏, 王芋丹, 等. 新疆地区腹泻仔猪源大肠杆菌的分群、血清型鉴定及耐药性分析[J]. 畜牧兽医学报, 2023, 54(1):414-420.
|
| [17] |
YIGIN A. Antimicrobial resistance and extended-spectrum beta-lactamase (ESBL) genes in E. coli isolated from equine fecal samples in Turkey[J]. Journal of Equine Veterinary Science, 2021, 101:103461.
doi: 10.1016/j.jevs.2021.103461
|
| [18] |
RESHADI P, HEYDARI F, GHANBARPOUR R, et al. Molecular characterization and antimicrobial resistance of potentially human-pathogenic Escherichia coli strains isolated from riding horses[J]. BMC Veterinary Research, 2021, 17(1):131.
doi: 10.1186/s12917-021-02832-x
|