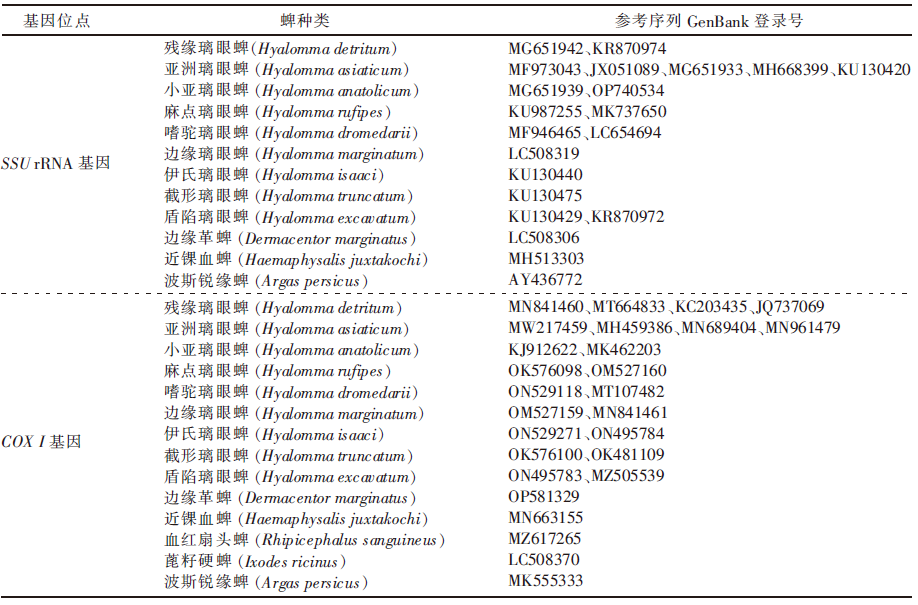
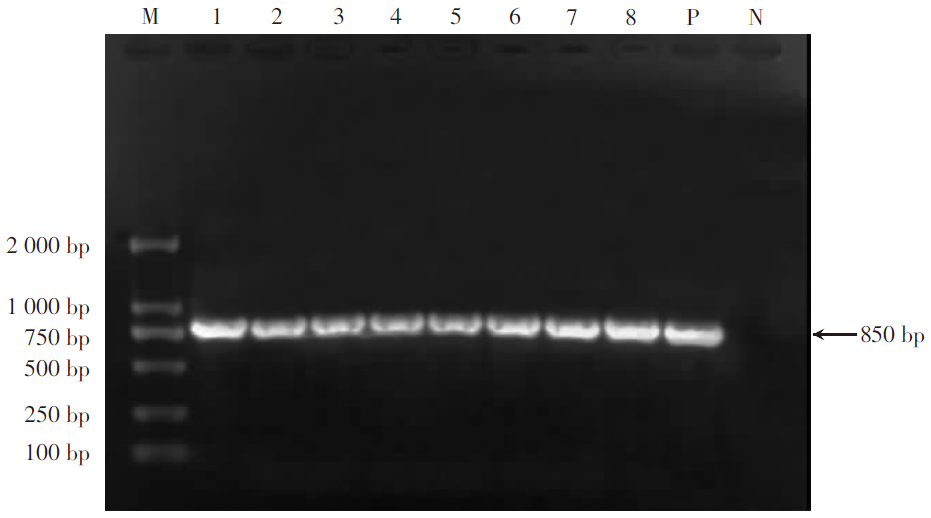
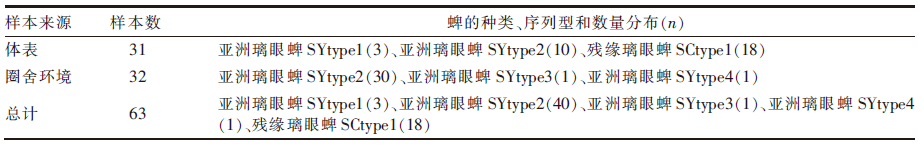
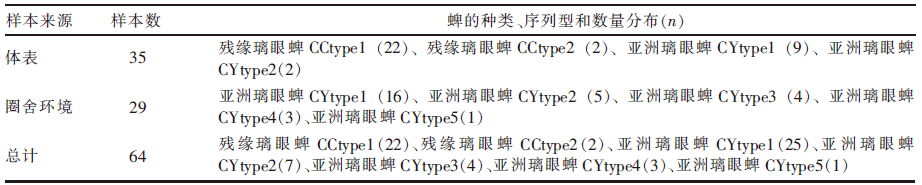
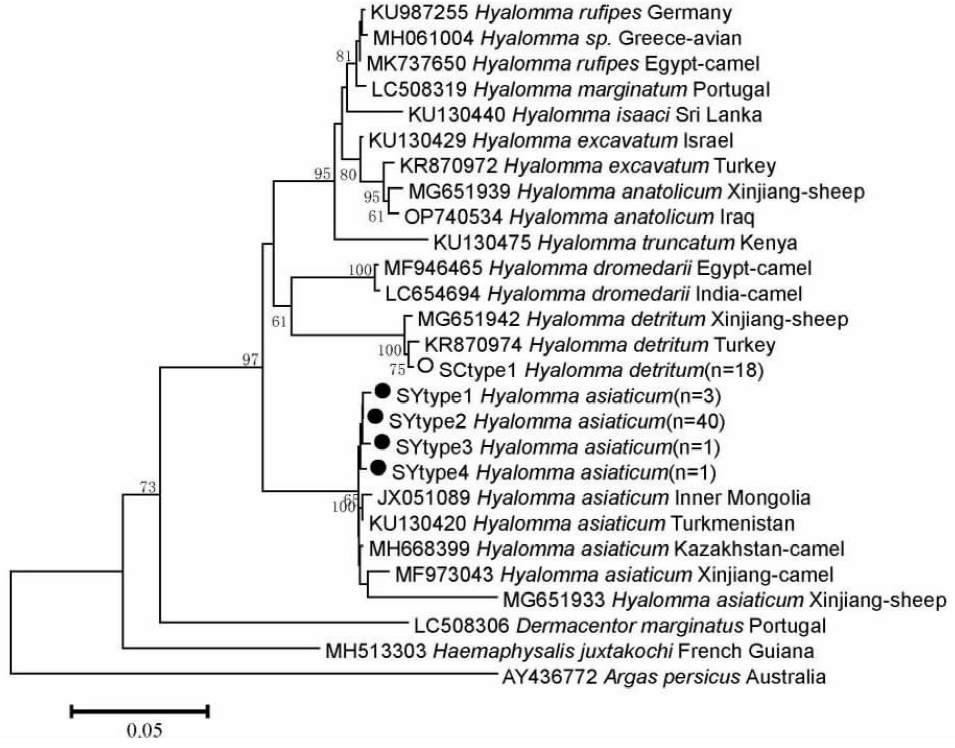
| [1] |
杨露, 朱长强, 艾乐乐, 等. 蜱携带重要人畜共患病毒研究概况[J]. 寄生虫与医学昆虫学报, 2018, 25(3):181-192.
|
| [2] |
张艳凯, 刘敬泽. 蜱类病原体和共生菌多样性及其作用[J]. 应用昆虫学报, 2019, 56(1):1-11.
|
| [3] |
于志军, 刘书广, 董娜, 等. 蜱类基础生物学研究进展[J]. 中国媒介生物学及控制杂志, 2018, 29(2):205-208.
doi: 10.11853/j.issn.1003.8280.2018.02.025
|
| [4] |
张桂林, 郑重, 孙响, 等. 新疆地区常见蜱种生态景观分布调查[J]. 中国媒介生物学及控制杂志, 2016, 27(5):432-435.
doi: 10.11853/j.issn.1003.8280.2016.05.003
|
| [5] |
于心, 叶瑞玉, 龚正达. 新疆蜱类志[M]. 乌鲁木齐: 新疆科技卫生出版社,1997.
|
| [6] |
杜景云, 牟路萌, 张璘, 等. 新疆边境地区伊宁县图兰扇头蜱形态学及分子生物学鉴定[J]. 中国媒介生物学及控制杂志, 2015, 26(3):271-274.
doi: 10.11853/j.issn.1003.4692.2015.03.013
|
| [7] |
SONG S, SHAO R F, ATWELL R, et al. Phylogenetic and phylogeographic relationships in Ixodes holocyclus and Ixodes cornuatus(Acari:Ixodidae)inferred from COX1 and ITS2 sequences[J]. International Journal for Parasitology, 2011, 41(8):871-880.
doi: 10.1016/j.ijpara.2011.03.008
|
| [8] |
沈华, 丁玉华, 徐安宏. 麋鹿长角血蜱病的诊治[J]. 草与畜杂志, 1996, 16(3):36.
|
| [9] |
孙大明, 朱明, 王桂宏, 等. 大量蜱寄生引致麋鹿贫血症病例报告[J]. 畜牧与兽医, 1998, 30(2):72-73.
|
| [10] |
王奇林, 蒋玉曦, 徐建平, 等. 塔里木马鹿体表寄生的软蜱种类及其携带病原的PCR鉴定[J]. 中国兽医科学, 2022, 52(3):339-343.
|
| [11] |
RUIZ-FONS F, GILBERT L. The role of deer as vehicles to move ticks,Ixodes ricinus,between contrasting habitats[J]. International Journal for Parasitology, 2010, 40(9):1013-1020.
doi: 10.1016/j.ijpara.2010.02.006
|
| [12] |
OLAFSON P U, BUCKMEIER B G, MAY M A, et al. Molecular screening for rickettsial bacteria and piroplasms in ixodid ticks surveyed from white-tailed deer(Odocoileus virginianus)and nilgai antelope(Boselaphus tragocamelus)in southern Texas[J]. International Journal for Parasitology:Parasites and Wildlife, 2020, 13:252-260.
doi: 10.1016/j.ijppaw.2020.11.002
|
| [13] |
SEO M G, KWON O D, KWAK D. Molecular detection of Rickettsia raoultii,Rickettsia tamurae,and associated pathogens from ticks parasitizing water deer(Hydropotes inermis argyropus)in South Korea[J]. Ticks and Tick-borne Diseases, 2021, 12(4):101712.
doi: 10.1016/j.ttbdis.2021.101712
|
| [14] |
SMALLEY R I V, ZAFAR H, LAND J, et al. Detection of Borrelia miyamotoi and powassan virus lineage Ⅱ(deer tick virus)from Odocoileus virginianus harvested Ixodes scapularis in Oklahoma[J]. Vector Borne and Zoonotic Diseases, 2022, 22(4):209-216.
doi: 10.1089/vbz.2021.0057
|
| [15] |
刘晓明, 张桂林, 赵焱, 等. 新疆尉犁荒漠地区蜱中主要蜱传疾病病原检测[J]. 中国媒介生物学及控制杂志, 2012, 23(6):496-498.
|
| [16] |
马博, 唐莉娟, 刘丹, 等. 巴州部分地区羊体表蜱类调查及防控措施[J]. 黑龙江畜牧兽医, 2021(3):99-102.
|
| [17] |
杜兰兰. 巴州牦牛三个试验点蜱传牛环形泰勒虫病的流行病学调查[D]. 乌鲁木齐: 新疆农业大学, 2018.
|
| [18] |
FILIPE D, PARREIRA R, PEREIRA A, et al. Preliminary comparative analysis of the resolving power of COX1 and 16S rDNA as molecular markers for the identification of ticks from Portugal[J]. Veterinary Parasitology:Regional Studies and Reports, 2021, 24:100551.
doi: 10.1016/j.vprsr.2021.100551
|
| [19] |
王云冲, 黄天鹏, 格日勒图. 内蒙古额济纳旗亚洲璃眼蜱的鉴定及其基因多态性分析[J]. 中国兽医学报, 2020, 40(12):2337-2341,2373.
|
| [20] |
CHITIMIA-DOBLER L, NAVA S, BESTEHORN M, et al. First detection of Hyalomma rufipes in Germany[J]. Ticks and Tick-borne Diseases, 2016, 7(6):1135-1138.
doi: 10.1016/j.ttbdis.2016.08.008
|
| [21] |
TELFORD S R. Deer reduction is a cornerstone of integrated deer tick management[J]. Journal of Integrated Pest Management, 2017, 8(1):25.
|